The molecular structure of Cerulenin
Click on the image above to interact with the 3D model of the Cerulenin structure
Cerulenin
Discovery (1,2), producing organism(1,2) and structure (3,4)
Cerulenin was originally isolated from the culture broth of the fungal strain KF-140 as an antifungal antibiotic with a structure of (2R,3S)-2,3-epoxy-4-oxo-7,10-trans, transdodecadienoylamide. The first paper on the total synthesis of cerulenin was reported by Boeckman et al (5).
Physical data (1)
White powder. C12H17NO3; MW 223.12. Sol. in EtOH, acetone, benzene. Slightly sol. in H2O. Practically insol. in petroleum ether.
Biological activity (2,6-10)
1) Inhibition of fatty acid synthesis (6-8)
Cerulenin is a potent inhibitor of fatty acid synthase (FAS). It inhibits all known types of
FASs; both multifunctional enzyme complexes (Type I) (from yeast, rat liver, mammalian
cells, and certain bacteria) and unassociated enzymes (Type II) (from most bacteria, and
higher plants).
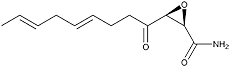
Cerulenin
2) Inhibition of polyketide synthesis (9,10)
Cerulenin blocks the synthesis of polyketides in a wide variety of organisms, including
actinomycetes, fungi and higher plants. In addition, cerulenin is suggested to inhibit the
condensation step in polyketide synthesis as well as fatty acid synthesis.
3) Antimicrobial activity (2)
Cerulenin has a wide range of antimicrobial activity. Most noteably, the
drug significantly inhibits growth of yeast-like fungi, such as Candida, Saccharomyces and Cryptococcus.
Mode of action (7,8, 11-19)
1) Inhibition of the condensing enzyme of FAS (7,8,11–13)
The inhibition of FAS activity by cerulenin is based on its covalent binding to the cysteine
residue (SpH) in the condensation reaction domain. Cerulenin was initially thought to take
a lactam form in order to react with the SpH (12). However X-ray crystallographic analysis
of the cerulenin-condensing enzyme complex revealed that cerulenin itself is covalently
attached in a hydrophobic cavity of the active site (13).
2) Mechanism of cerulenin-resistance in a cerulenin producer (14-18) and mutant yeast (17)
The cerulenin-producing fungus Acremonium (Cephalosporium) caerulens KF-140 was
resistant to cerulenin. The mechanism of self-resistance revealed that cerulenin cannot
bind to the significantly altered condensation reaction domain of the producer FAS(13-15).
In a cerulenin resistant mutant (KNCR-1) (a 30-fold increase) of Saccharomyces cerevisiae,
the GGT codon encoding Gly-1257 of FAS 2, (the gene encoding the α subunit of the
fatty acid synthase) was changed to AGT, resulting in the codon for Ser(16). As predicted,
Gly-1257 is incorporated in the hydrophobic cavity of the active site to fit cerulenin(18).
Therefore, the change causes steric hindrance to cerulenin binding, leading to cerulenin
resistance in mutant yeast.
Total synthesis (20)
The total synthesis of cerulenin has been reported by many groups.
---
Cerulenin is commercially available as an biochemical reagent in the field of lipid research.
The antibiotic was used for production of hybrid compounds
---
Synthetic derivative CM-55
1) Synthesis and structure (21)
CM-55 was synthesized as an analog of antilipogenic antibiotic cerulenin.
2) Biological activity (21-23)
i) Antimicrobial activity (21,22)
CM-55 shows antifungal and antimicrobial activity with rather low MIC values (6.25-12.5 µg/ml)(21). The mode of action appears to be attributed to inhibition of acetyl-
CoA by incorporation into the non-saponifiable fraction (22).
ii) CM-55 was found to inhibit epithelial transport of sodium ions across the toad urinary bladder and to inhibit hydro-osmotic response of the bladder to vasopressin after 2 hours of pretreatment with CM-55 (23).
References
1. [17] Y. Sano et al., J. Antibiot., 20, 344–348 (1967)
2. [119] S. Ōmura, Bacteriol. Rev., 40, 681–697 (1976)
3. [18] S. Ōmura et al., J. Antibiot., 20, 349–354 (1967)
4. [71] B. H., Arison and S. Ōmura, J. Antibiot., 27, 28–30 (1974)
5. R. K. Jr. Boeckman et al., J. Am. Chem. Soc., 99, 2905 (1977)
6. [219] S. Ōmura, Methods in Enzymol., 72, 520–532 (1981)
7. [244] A. Kawaguchi et al., J. Biochem., 92, 7–12 (1982)
8. [51] D. Vance et al., Biochem. Biophys. Res. Commun., 48, 649–656 (1972)
9. [70] S. Ōmura and H. Takeshima, J. Biochem., 75, 193–195 (1974)
10. [102] H. Ohno et al., J. Biochem., 78, 1149–1152 (1975)
11. [69] G. D’Agnolo et al., Biochim. Biophys. Acta., 326, 155–166 (1973)
12. [414] H. Funabashi et al., J. Biochem., 105, 751–755 (1989)
13. [372] H. Tomoda et al., J. Antibiot., 40, 1457–1460 (1987)
14. [173] A. Kawaguchi et al., Arch. Biochem. Biophys., 197, 30–35 (1979)
15. [284] H. Tomoda et al., J. Biochem., 95, 1705–1712 (1984)
16. [285] H. Tomoda et al., J. Biochem., 95, 1713–1723 (1984)
17. [541] J. Inokoshi et al., Mol. Gen. Genet., 244, 90–96 (1994)
18. M. Moche et al., J. Biol. Chem., 274, 6031 (1999)
19. [816] H. Tomoda et al., Proc. Japan Acad., 78, 217-240 (2002)
20. [243] M. Tishler et al., J. Org. Chem., 47, 1221-1228 (1982)
21. [82] S. Ōmura et al., Antimicrob. Agents Chemother., 6, 207-215 (1974)
22. [84] T. Ohno et al., Antimicrob. Agents Chemother., 6, 387-392 (1974)
23. [79] T. Ohno et al., J. Membrane Biol., 18, 295-304 (1974)


